
However, in large networks this will over-stress the computer and become sluggish in the interface. If the chart is in its interactive mode, you can see the main graph view changing its selection as the chart changes. To clear the selection, click outside of the range. To edit an existing selection, drag in the middle of the selection to move the entire selection, or on either edge of the selection to edit just the start or end of the range. You should see a color change in the background of the data. To select a range within a histogram, click and drag left or right. The structure of the c圜hart window has a header with common functions and settings, the content of the chart, and a footer with the selections status and the controls to set the axes. To create a histogram using c圜hart, open the right-click on the header of a numeric column in the Node or Edge Table, and select the command Plot Histogram… This can be particularly useful for finding pockets of the data that express similar ranges, such as positively and negatively expressed genes.
It shows the user where the most common values are and whether the values are distributed uniformly (flat line), normally (the bell curve) or have strong modes (hills and valleys).

The file format is simply oneĪ histogram shows the distribution of a variable in bins over a range. Select → Nodes → From ID List File… selects nodes based on node Select → Mouse Drag Selects includes the same options for Selection Modeįor mouse click or drag-selection as the current Selection Mode in the Network Select all (nodes, edges and annotations)ĭeselect all (nodes, edges and annotations) Regardless of which Selection Mode for mouse click or drag-selection. Most options are fairlyĪ set of keyboard shortcuts for selecting nodes, edgesĪnd annotations are also available. Mechanisms for selecting nodes and edges. The Select → Nodes and Select → Edges menus provide several To search for column values that contain special characters you need toĮscape those characters using a \. Letters and numbers currently do not work when searching a specificĬolumn. If you don’t specify a particular column, all columnsĬolumns with names that contain spaces, quotes, or characters other than That have a COMMON column value that starts with STE, useĬommon:ste*. To search a specific column, you can prefix your search term with theĬolumn name followed by a. Ste? would match STE2 but would not match STE12.
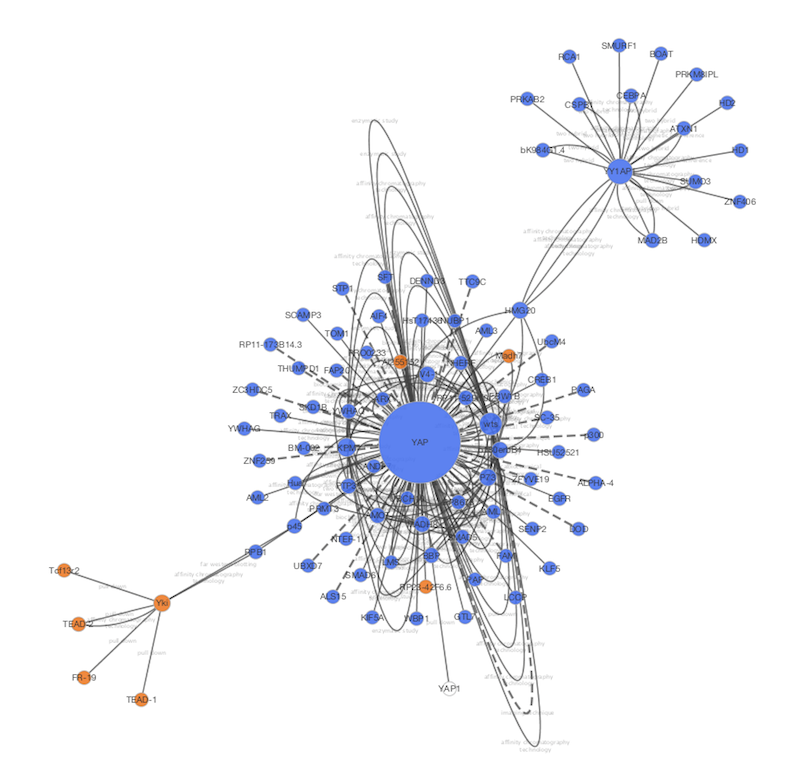
Zero or more characters, while ? matches exactly one character. The * is a wildcard character that matches For example, to select nodes or edges with aĬolumn value that starts with STE, type ste* in the search bar. These results demonstrate the utility of SAFE for examining biological networks and understanding their functional organization.You can search for nodes and edges by column value directly throughĬytoscape’s tool bar. Integration of genetic interaction and chemical genomics data using SAFE revealed a link between vesicle-mediate transport and resistance to the anti-cancer drug bortezomib. SAFE annotations of the genetic network matched manually derived annotations, while taking less than 1% of the time, and proved robust to noise and sensitive to biological signal. I applied SAFE to annotate the Saccharomyces cerevisiae genetic interaction similarity network and protein-protein interaction network with gene ontology terms. SAFE visualizes the network in 2D space and measures the continuous distribution of functional enrichment across local neighborhoods, producing a list of the associated functions and a map of their relative positioning. Here, I describe spatial analysis of functional enrichment (SAFE), a systematic method for annotating biological networks and examining their functional organization. Large-scale biological networks represent relationships between genes, but our understanding of how networks are functionally organized is limited.


 0 kommentar(er)
0 kommentar(er)
